Updated covariance models of microRNA families
To address challenges manual annotation of microRNAs, MirMachine was developed in 2023. The initial version, based on MirGeneDB 2.1, uses covariance models (CMs) of 508 conserved microRNA families to detect their presence in a species’ genome, even without small RNA data. CMs are particularly effective in detecting homologous RNA sequences due to their ability to capture both the primary sequence information and the secondary structural constraints of microRNAs. Despite its effectiveness, MirMachine’s covariance models initially favored well-sampled clade. To mitigate this, the models were retrained using the expanded species set from MirGeneDB 3.0, which reduces false negatives and enhances sequence diversity tolerance.
Here our models are separated into three groups: combined models refer to CMs trained using all MirGeneDB, deuterostome models refer to CMs trained using only deuterostome species available in MirGeneDB, and lastly protostomes models refer to CMs trained using only protostomes species available in MirGeneDB. Our updated covariance models are now hosted here (see right site) and will feature in a future version of MirMachine. However, they can already be used with the Infernal’s (http://eddylab.org/infernal/) cmsearch function.
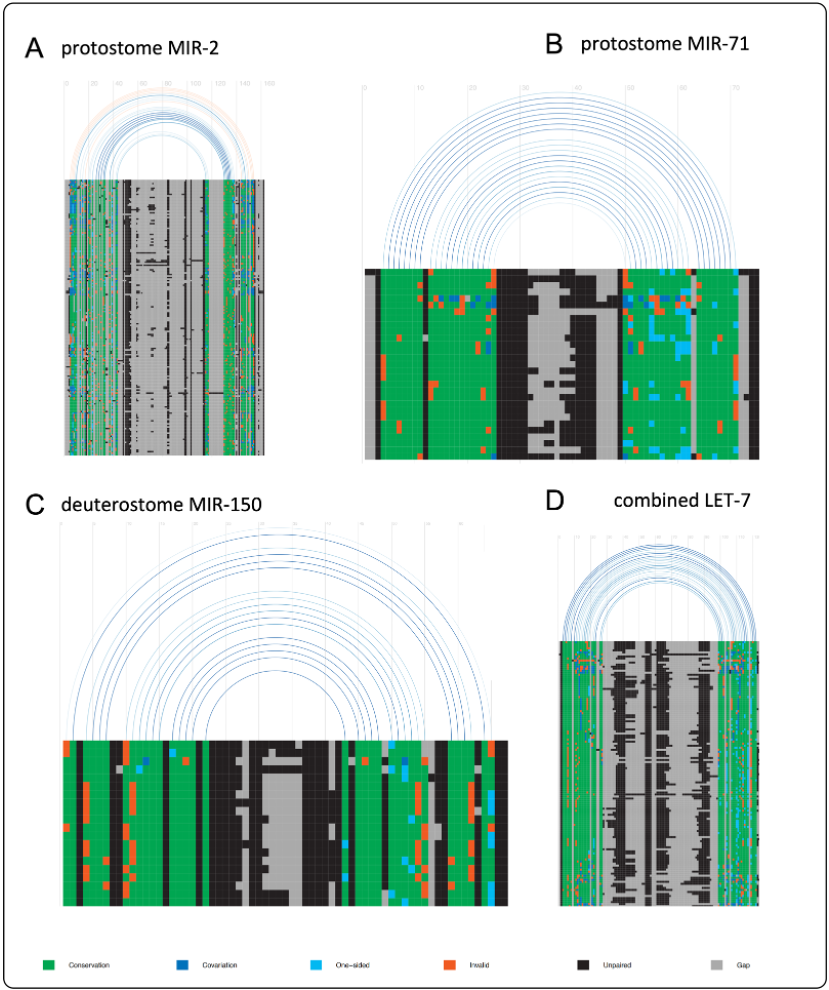
Graphical representations of CMs of selected microRNA families.






